Essential Protein Chemistry
- Primary structure of proteins
- Secondary structure of proteins
- Tertiary structure of proteins
- The importance of protein bonding interactions
- Quaternary structure of proteins
1 Primary structure of proteins
Functional Roles of Proteins
| Hormone | Regulates body processes |
|---|---|
| Enzyme | Biological catalyst |
| Protective | Combats infection (e.g. antibody) |
| Storage | Nutrient storage (e.g. casein) |
| Structure | Forms part of an organism's structure |
| Transport | Movement of substances in the body |
Proteins are polymeric amino acids of high molecular weight, arbitrarily 10,000 Da or more, which fulfil important roles in an organism's life cycle. Some useful definitions of these roles are shown in the box on the right.
Proteins can be classified with reference to any of several distinguishing features (molecular weight, occurrence, solubility, etc.). One distinction is between simple and conjugated proteins: Simple proteins are composed entirely of amino acids; conjugated proteins have a non-amino acid component (called a prosthetic group).
The building blocks of proteins are approximately twenty naturally occurring alpha-amino acids (for structures, see Appendix 3: Proteinogenic Amino Acids). These can form amides by the condensation of the amine group of one amino acid with the carboxylic acid group of another. When the new amide bond involves two of the natural amino acids, it is referred to as a peptide bond.

A peptide 'dimer' made from two amino acids still contains a free carboxyl group and a free amino group, so it can form more peptide bonds through either of these groups. The products are classified according to the number of amino acid units involved. The structure below is a pentapeptide; there are less specific names such as oligopeptides ('a few') and polypeptides ('many'). A polypeptide can also be described as an 'n-mer' if n amino acids are connected.

Primary structure
The convention for drawing peptides is to put the N-terminus on the left. The amino acid at the left-hand end is referred to as the N-terminal amino acid, and the amino acid at the right-hand end is referred to as the C-terminal amino acid. Three-letter codes are very useful for describing polypeptides without having to draw them. For example, the pentapeptide met-enkephalin can be written as H-Tyr-Gly-Gly-Phe-Met-OH.
In general, when polymerising n different amino acids (using each once) there are n! possible sequences of product. It is this sequence of amino acids in a polypeptide chain that is referred to as its primary structure. An octapeptide has 40,320 possible primary structures. Nature clearly has many options for the construction of proteins from different permutations of the 20-odd amino acids that are routinely available.
Because the α-amino acids are single enantiomers, the polypeptides and proteins that are formed from them are also chiral (handed) molecules. Proteins are not only long chains, but fold up into complex 3-D structures that include lengths of randomly coiled chain along with regions of ordered structure. The nature of these is discussed in the following sections.
2 Secondary structure of proteins
The term secondary structure refers to the geometry of a polypeptide's N-C-C backbone, which can be randomly coiled or may be ordered in special ways. The geometry is strongly influenced by two factors:
The stereochemistry of the amide link. A key structural feature of polypeptides and proteins is the planarity of an amide bond, in which the N-atom has sp2 (trigonal planar) geometry. The peptide bond thus has significant double-bond character (bond length 1.32 Å) which inhibits rotation about the N–CO bond: in this respect amides resemble alkenes. This has important consequences in the context of protein secondary and tertiary structure.

As is clearly indicated in the resonance picture above, the carbonyl oxygen and the amide hydrogen have respective partial negative and positive charges (Oδ− and Hδ+), leading to the possibility of intra- and interchain hydrogen bonding. Hydrogen bonding is a key aspect of protein structure, and the way that it is organised can control secondary structure.
The α-helix results from coiling of the NCC backbone with the formation of H-bonds between the C=O of residue (n) and the NH of residue (n+4). This arrangement readily accommodates large 'R' groups since they all point outwards. Every C=O and NH group is involved in hydrogen bonding. Other helices can occur, although they are less common. Examples are the 3(10)-helix and the π-helix.
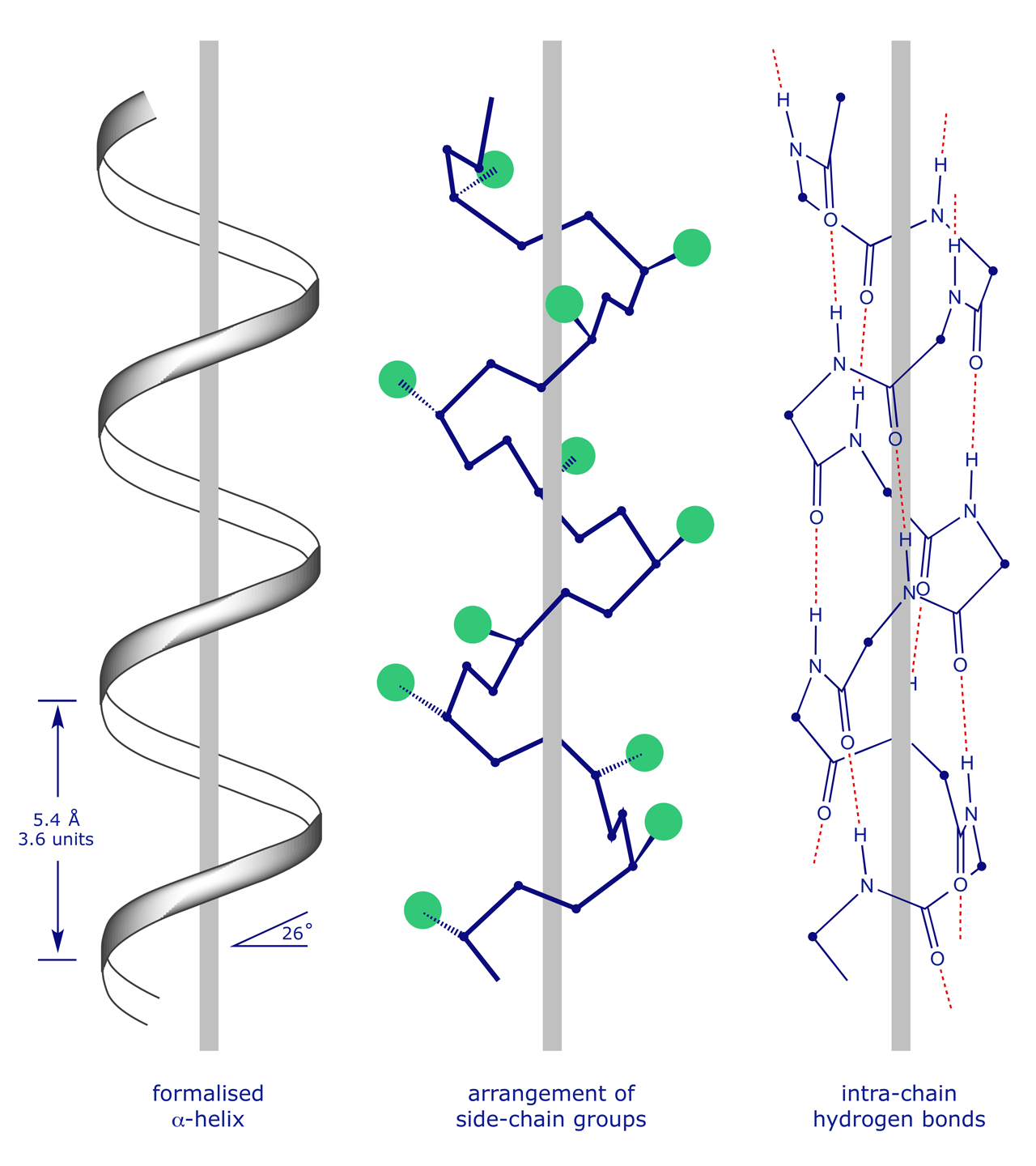
The β-sheet has layering of protein chains, one on top of the other, again with hydrogen bonds between the chains. Side-chain 'R' groups are at right-angles to the sheets, minimising steric interactions. The chains can run in opposite directions (antiparallel) or the same direction (parallel). In 'cartoon' structures (discussed later), the direction is indicated by an arrow.

The β-turn changes the direction of a protein NCC backbone so that, effectively, it doubles back on itself. This is an important feature of protein folding, which gives a more compact shape. The β-turn is stabilised by a hydrogen bond between the first and third peptides. The amino acid proline is frequently found at bends in a protein backbone.

3 Tertiary structure of proteins
Many physiologically active proteins do not have the regular structure of the helix or sheet, but rely for their properties on their tertiary structure, which can be defined as modifications in the 3-D structure due to interactions between the side-chains of the amino acids. The chief interactions are S–S covalent links, polar/ionic (electrostatic) attractions, H-bonding and van der Waals forces, which are explained below. An illustration is provided by myoglobin, a protein that mediates O2 transport in muscle tissue. The secondary structure (backbone) of myoglobin is 70% α-helix, but its functionality relies on its overall hemispherical shape — the tertiary structure. This features polar groups on the outside and non-polar groups on the inside, making it water-soluble. Proline residues occur at the bends, as do some amino acids which do not readily form α-helical coils, e.g. isoleucine and serine.
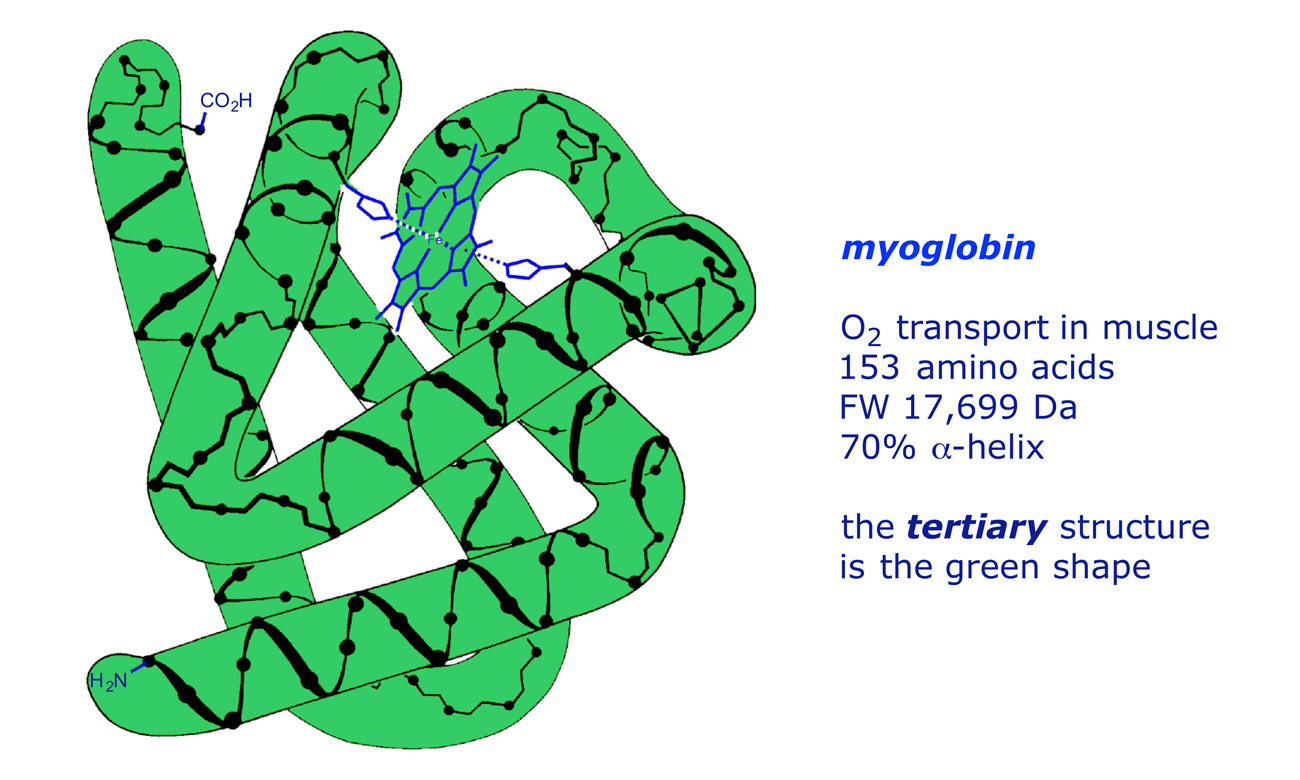
The bonding that can hold a protein in its tertiary structure, i.e. its globular shape, includes the following:
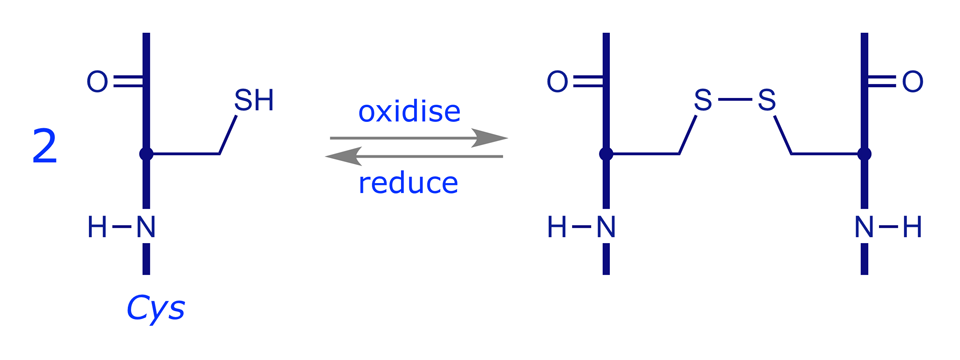
Disulfide Bridging. Two cysteine residues can be oxidised at their thiol (–SH) groups so as to form a disulfide bridge (–S–S–). Reduction reverses this process. The bridges can link two different chains or different parts of the same chain.
Polar/Ionic Interactions. At physiological pH (7) some side-chain groups are protonated (cationic) while others are deprotonated (anionic). Interaction of these can give an ionic (electrostatic) bond or 'salt bridge' which again may be interchain or intrachain.

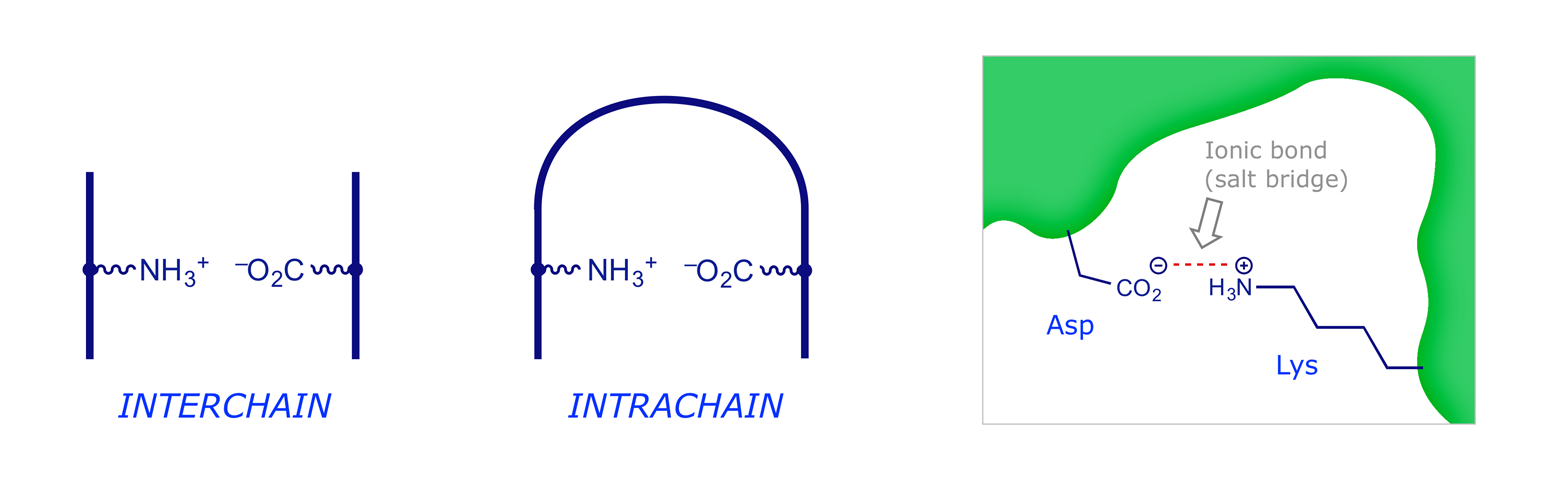
Hydrogen Bonding. Due to electronegativity differences, the N–H bonds in peptides are polarised with Hδ+ and the carbonyls are polarised with Oδ−. Bonding between these gives weak but numerous hydrogen bonds, which again may be interchain or intrachain.
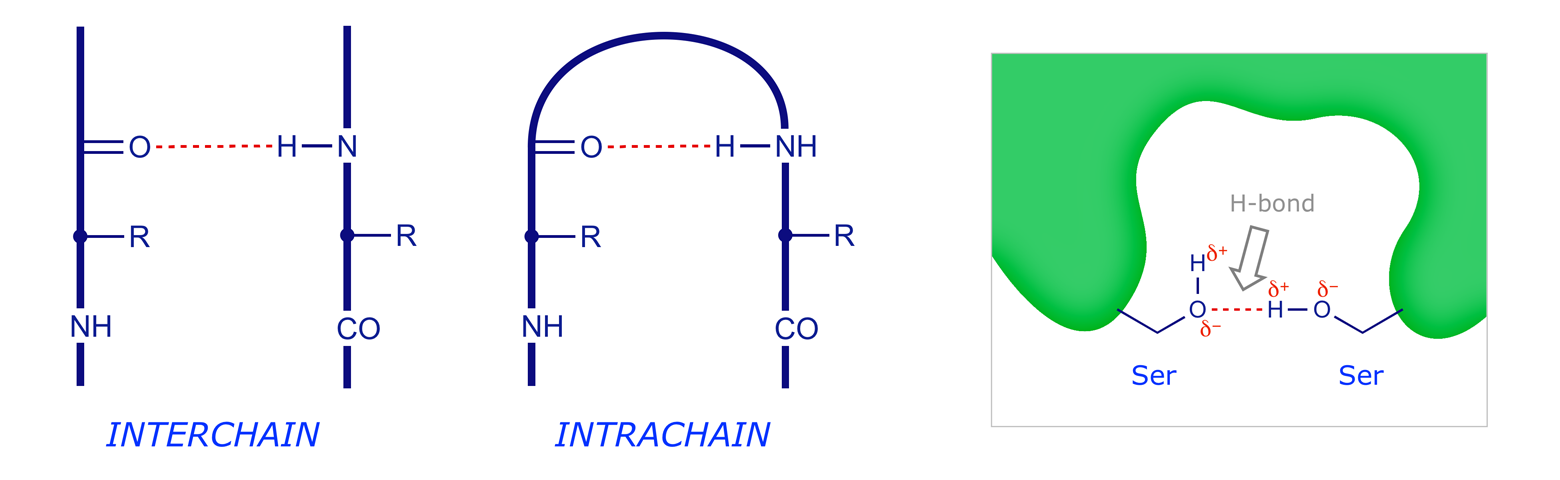
Van der Waals Interactions. These are very weak, and occur between hydrophobic (non-polar) side-chains, which associate with each other rather than the aqueous (i.e. hydrophilic) medium. The amino acids involved are those with non-polar side-chains, e.g. Phe, Tyr, Gly, Ala, Val, Leu and Ileu.

4 The importance of protein bonding interactions
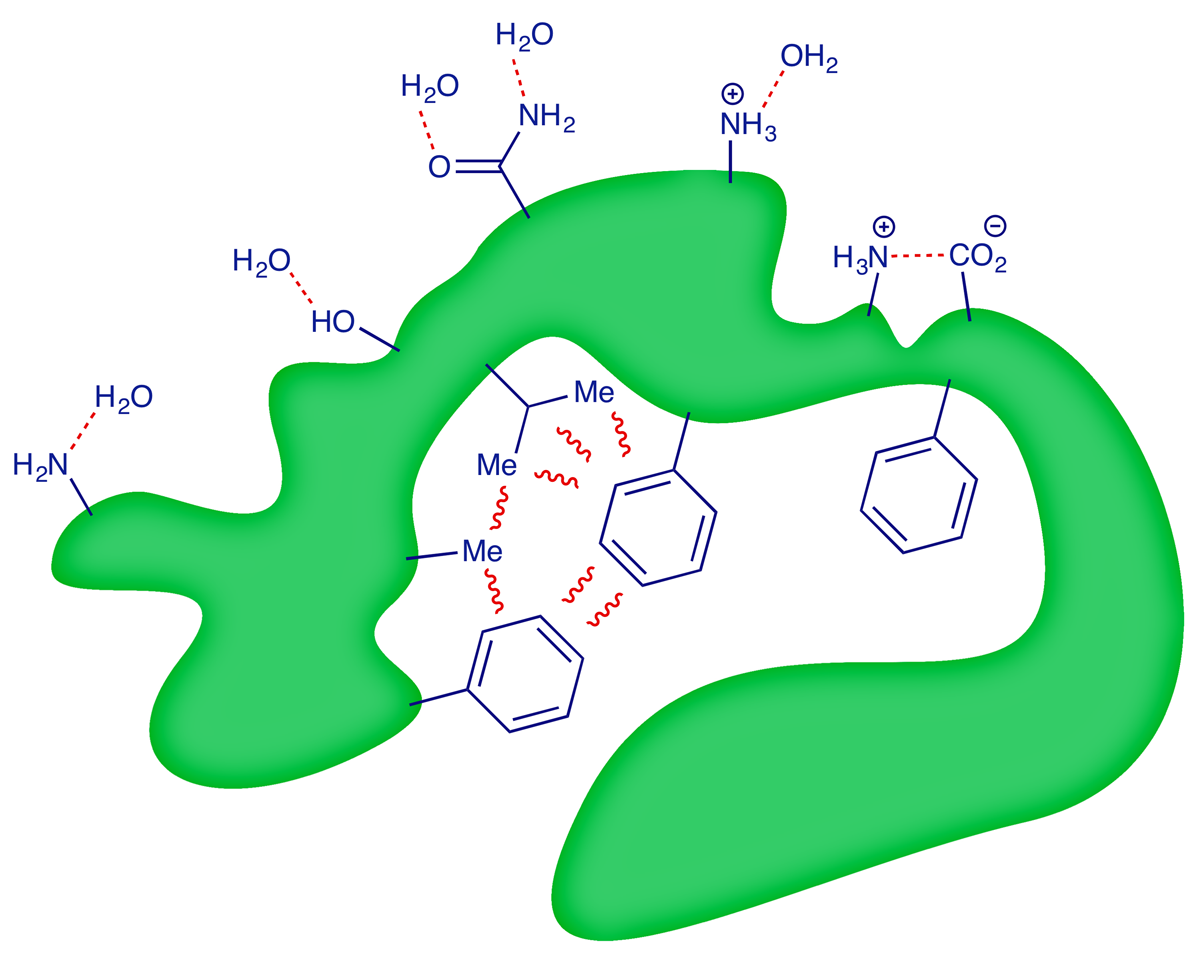
From the point of view of functionality, the most influential contributions to a protein's tertiary structure are made by the 'weaker' forces, hydrogen bonding and van der Waals interactions. To begin with, more opportunities exist for these to be influential: only a few amino acids can participate in ionic bonding and only one (Cys) can offer S–S bonds. More importantly, because proteins 'operate' in water, the most stable tertiary structure will have the polar groups on the outside and the hydrophobic groups on the inside, where they avoid the water but interact with each other at close quarters. It is the resulting 'shape' of the protein interior that ultimately determines the tertiary structure. This is exploited by drug designers, who seek small molecules that will be drawn to coordination sites on the interior of the protein, changing its shape and hence its activity.
The cartoon below summarises how interactions and repulsions between the side-chain groups of the amino acids in a protein, and the solvent water, affect its tertiary structure. The protein will undergo conformational changes until the sum of the interactions is minimised.
5 Quaternary structure of proteins
Some globular proteins are oligomeric, i.e. contain two or more separate polypeptide chains. These have a quaternary structure which is defined as the way in which the individual folded protein chains fit together. Hemoglobin has an overall spherical shape but was found, after 25 years of study by M. F. Perutz and coworkers, to consist of two α chains (each 141 amino acids) and two β chains (each 146 amino acids). The hemoglobin molecule has four reactive heme groups groups, each partially buried in a pocket lined with non-polar R groups. This is important: the absence of water inhibits the oxidation of the iron from +2 to +3 when it is bound to oxygen.

Van der Waals interactions between exterior hydrophobic parts of the contributing sub-units help to maintain the quaternary structure of a protein aggregate in water.

